Overriding materials
Since version >= 3.75
Purpose: to show usage of override: directive for replacing materials in a voxelized region using ROIs.
We create a synthetic CT consisting of a spherical skull filled with brain material. The skull is a spherical wall of cortical bone 1 cm thick. The skull is surrounded by air. We nicknamed this setup as the Mental Ball.
The input file looks like:
# create a "Mental Ball" and deliver proton beam
region: phantom; CTscan = CT.mha ; pivot=[0.5,0.5,0]; O=[0,0,0]; score = [dose]; lWriteDensity=t
override: phantom HU-1000 outside Sphere8cm.mha
override: phantom HU+1376 inside Sphere8cm.mha
override: phantom HU+32 inside Sphere7cm.mha
lUseInternalHU2Mat=t
pb: 1 0 ; Xsec = disc ; FWHM = 4 ; particle = p ; E = 100; N=1e8
nprim = 1e5
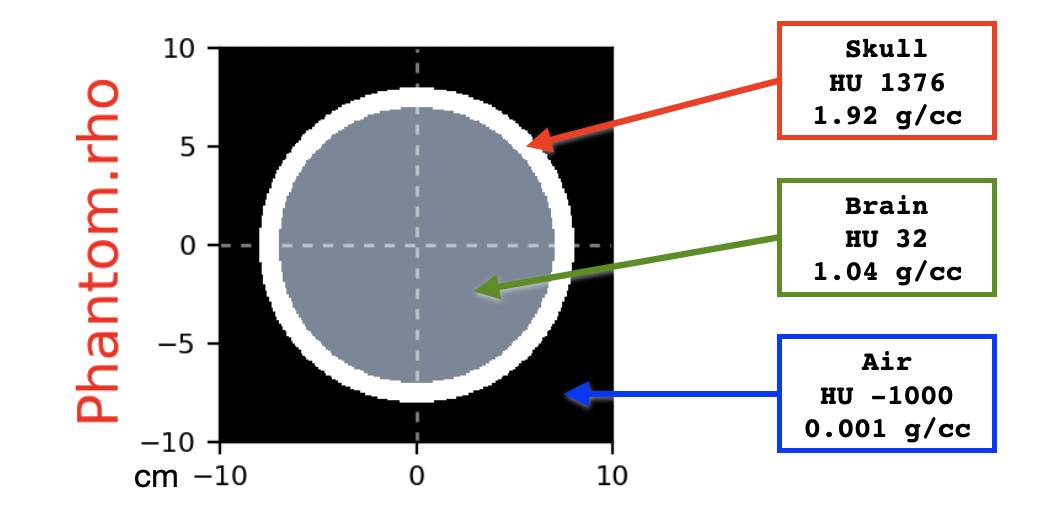
Density map of the Mental Ball
Explanation of the procedure:
load a CT scan into the Phantom region
set air (HU -1000) outside a sphere of radius 8 cm
set cortical bone (HU +1376) inside the same sphere
replace with brain (HU +32) a sphere of radius 7 cm
Delivering the beam, we obtain the following dose map
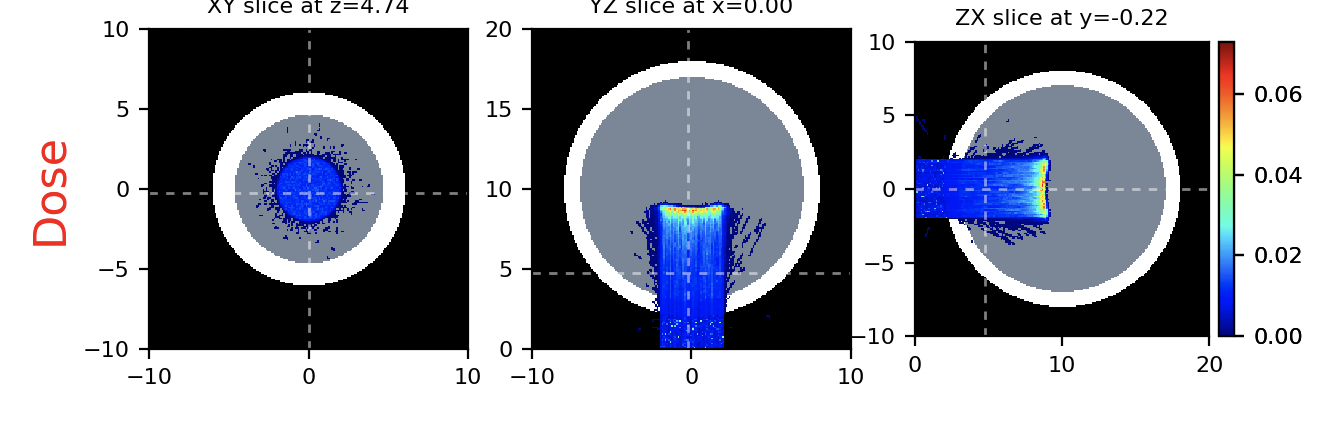
Important
the override: rules are applied in the order they are found in the input file
Hint
It is possible to mix standard and HU materials. For instance:
override: phantom air outside Sphere8cm.mha
override: phantom HU1300 inside Sphere8cm.mha
override: phantom HU32 inside Sphere7cm.mha
or
override: phantom vacuum outside Sphere8cm.mha
override: phantom Al inside Sphere8cm.mha
override: phantom water inside Sphere7cm.mha
Python script for generating the synthetic CT and the ROIs for the `Mental Ball`
The following python script shows how to generate a synthetic CT and two ROIs. The ROIs are two filled spheres with a radius of 8 and 7 cm respectively.
import os
import numpy as np
import SimpleITK as sitk
########################################################################
Lx = 200 ; Ly = 200 ; Lz = 200 # extent (mm)
nx = 200 ; ny = 200 ; nz = 200 # voxels
hx = Lx/nx ; hy = Ly/ny ; hz = Lz/nz # spacing
print('extent',Lx,Ly,Lz,'mm')
print('voxels',nx,ny,nz,'#')
print('spacing',hx,hy,hz,'mm')
# center voxel coords
x = np.linspace(-Lx/2,+Lx/2,nx,endpoint=False)+hx/2
y = np.linspace(-Ly/2,+Ly/2,ny,endpoint=False)+hy/2
z = np.linspace(-Lz/2,+Lz/2,nz,endpoint=False)+hz/2
zv, yv, xv = np.meshgrid(z,y,x,indexing='ij')
# CT map corresponding to HU=0 value
CT = np.zeros((nz,ny,nx),dtype=np.int16)
img = sitk.GetImageFromArray(CT)
img.SetSpacing((hx,hy,hz))
img.SetOrigin((x[0],y[0],z[0]))
sitk.WriteImage(img,'CT.mha')
# ROI of a sphere with 8 cm radius
ROI = np.zeros_like(CT)
ROI[xv**2+yv**2+zv**2<80**2]=1
img = sitk.GetImageFromArray(ROI)
img.SetSpacing((hx,hy,hz))
img.SetOrigin((x[0],y[0],z[0]))
sitk.WriteImage(img,'Sphere8cm.mha')
# ROI of a sphere with 7 cm radius
ROI = np.zeros_like(CT)
ROI[xv**2+yv**2+zv**2<70**2]=1
img = sitk.GetImageFromArray(ROI)
img.SetSpacing((hx,hy,hz))
img.SetOrigin((x[0],y[0],z[0]))
sitk.WriteImage(img,'Sphere7cm.mha')