Radiobiology
FRED can compute the effective biological dose using several built-in RBE models for protons.
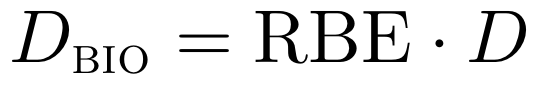
where the relative biological effectiveness (RBE) for proton beams is described using the linear-quadratic model

where (alpha/beta)x is the reference ratio for X-rays (see, for instance, Polster L. et al, Phys. Med. Biol. 60 (2015) 5053).
The reference values of alpha_X and beta_X can be defined in the input file (default values are shown hereafter):
RBE_alphaX = 0.0722
RBE_betaX = 0.0502
New in version 3.70.0
It is possible to prescribe a different value for
alpha_Xandbeta_Xfor each organ in a CT scan. You can do this by loading two maps for each region as in the following example. The values can be actually defined at the voxel level. Setting voxels to 0 will actually correspond to mask those part of the CT, and they will not be included in the final RBE calculation.region< ID=phantom CTscan='inp/CT_ExtROICrop_1.5x1.5x1.5.mhd' score=[Dose,RBE,LETd] alphaBetaX=[inp/myAlphaX.mhd,inp/myBetaX.mhd] region>
Note
The results of RBE models (e.g. maps of RBE, Dose_Bio, alpha and beta parameters) are saved in the directory out/RBE which is created only if RBE-based calculations are requested.
Constant RBE
The most simple assumption is to consider proton RBE constant with respect to the dose level delivered to the patient. Given the complexity of the topic, the constant RBE is still a working solution adopted in many treatment centres worldwide. The typical reference value is 1.1 for protons, namely proton dose is on average 10% more effective than the same dose delivered by photons.
- RBE_constant = (float)
if set, it activates the conversion of physical dose to biological dose using the given constant value. The corresponding dose map is saved in
out/RBE/DoseBio_Constant.mhd.
Variable RBE models are grouped into LETd-based models and Table-based models.
LETd-based models
The following section presents the implemented models with the default values that can be overridden in the input file.
Hint
You can use the built-in models described below. It is anyhow very simple to implement new RBE models as post-processing of FRED output. For inspiration, have a look at the python script in the examples that re-implements the McNamara model.
Carabe
lRBE_Carabe = t # activate model
# model parameters and default values:
RBE_Carabe_p1 = 0.843
RBE_Carabe_p2 = 0.154
RBE_Carabe_p3 = 2.686
RBE_Carabe_p4 = 1.09
RBE_Carabe_p5 = 0.006
Reference: Carabe-Fernandez A, Dale R G and Jones B, The incorporation of the concept of minimum RBE (RBEmin) into the linear-quadratic model and the potential for improved radiobiological analysis of high-LET treatments, Int. J. Radiat. Biol. 83 27–39 (2007)
Chen
lRBE_Chen = t # activate model
# model parameters and default values:
RBE_Chen_lambda1 = 0.0013
RBE_Chen_lambda2 = 0.045
RBE_Chen_alpha1 = 0.1
Reference: Chen Y and Ahmad S, Empirical model estimation of relative biological effectiveness for proton beam therapy, Radiat. Prot. Dosim. 149 116–23 (2012)
McNamara
lRBE_McNamara = t # activate model
# model parameters and default values:
RBE_McNamara_p0 = 0.99064
RBE_McNamara_p1 = 0.35605
RBE_McNamara_p2 = 1.1012
RBE_McNamara_p3 = -0.0038703
Reference: McNamara AL, Schuemann J, Paganetti H, A phenomenological relative biological effectiveness (RBE) model for proton therapy based on all published in vitro cell survival data. Phys Med Biol. 60 8399-416 (2015)
Wedenberg
lRBE_Wedenberg = t # activate model
# model parameters and default values:
RBE_Wedenberg_c1 = 0.434
Reference: Wedenberg M, Lind B K and Hardemark B, A model for the relative biological effectiveness of protons: the tissue specific parameter alpha/beta of photons is a predictor for the sensitivity to LET changes, Acta Oncol. 52 580–8 (2013)
Wilkens
lRBE_Wilkens = t # activate model
# model parameters and default values:
RBE_Wilkens_lambda = 0.02
RBE_Wilkens_alpha0 = 0.1
Reference: Wilkens J J and Oelfke U, A phenomenological model for the relative biological effectiveness in therapeutic proton beams, Phys. Med. Biol. 49 2811–25 (2004)
Note
The amount of information that is saved to disk for each requested RBE model is controlled by several flags.